Skills






Experience
AI Systems Research Intern, Los Alamos National Lab
Los Alamos, NM
- Built a modular multi-agent AI workflow with RAG capabilities to automate terabyte-scale simulation data analysis, in collaboration with Argonne National Lab.
- Integrated LangChain and LangGraph for schema-aware database querying and intelligent task delegation to support plug-and-play scientific pipelines.
- Evaluated modularity, scalability, and interpretability in agentic system design to inform future scientific AI infrastructure at LANL.

Bioinformatics Research Co-op, Moderna Therapeutics
Cambridge, MA
- Developed a PyTorch-based transformer model and graph neural network model for mRNA analysis
- Discovered a novel method for altering normally inflexible mRNA structural properties using graph features.
- Engineered several production-level features and improvements to the current Moderna computational pipeline.
- Presented comprehensive reviews and project insights to executive leadership at Moderna, including the VP and directors of computational science.

Adjunct Professor in Mathematics
Brooklyn, NY
- Aided in the development of a web-based interactive homework assignment and grading system, used in all math classes throughout the college.
- Taught several college level mathematics courses including pre-calculus, calculus 1, and calculus 2.
- Utilized a mix of lecture-style and activity-style teaching strategies to maximize participation and understanding of difficult topics.

Publications
InferA: A Smart Assistant for Cosmological Ensemble Data
Workshops of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC Workshops ’25), St Louis, MO, USA, November 16–21. New York, NY: Association for Computing Machinery.
A provenance-aware multi-agent workflow that interprets natural language queries to automate data analysis and generate visualizations on terabyte-scale simulation datasets.
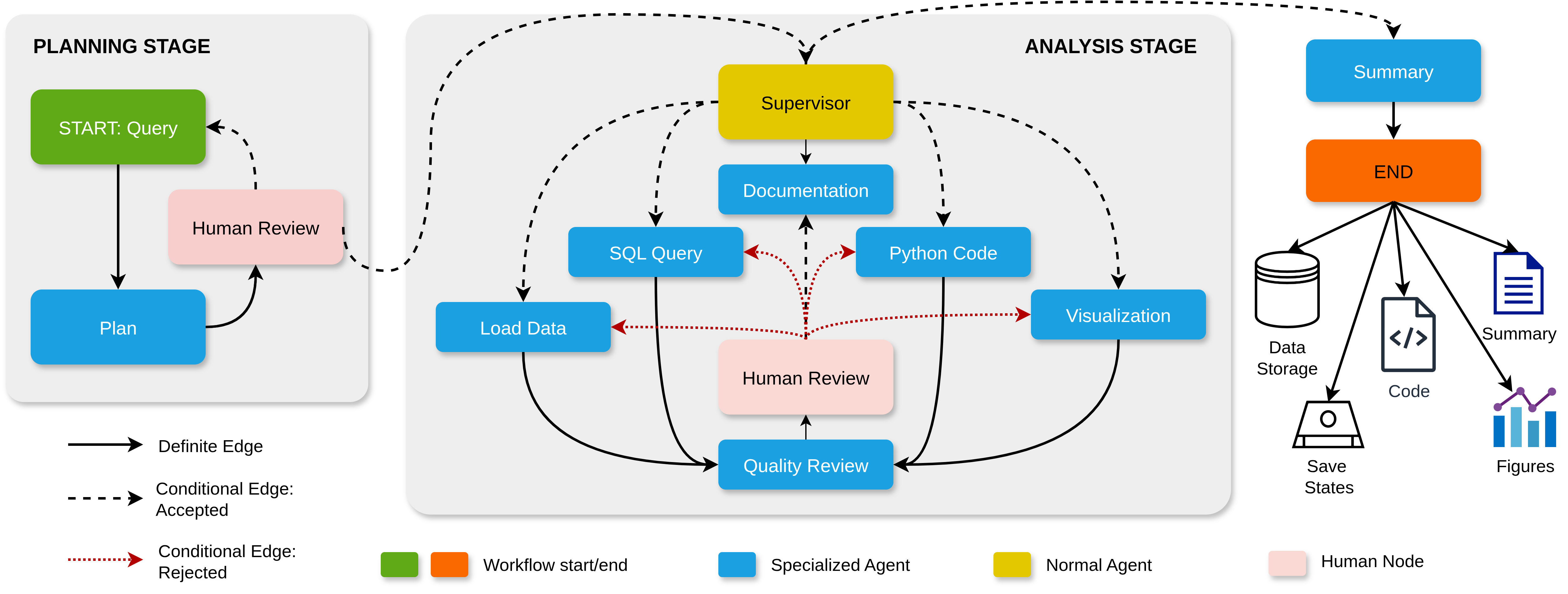
Automatic Explanation of Protein-Protein Binding Mechanism: A Preliminary Study
Computational Structural Bioinformatics Workshop (pp. 84-97). Cham: Springer Nature Switzerland, Mar 26, 2025
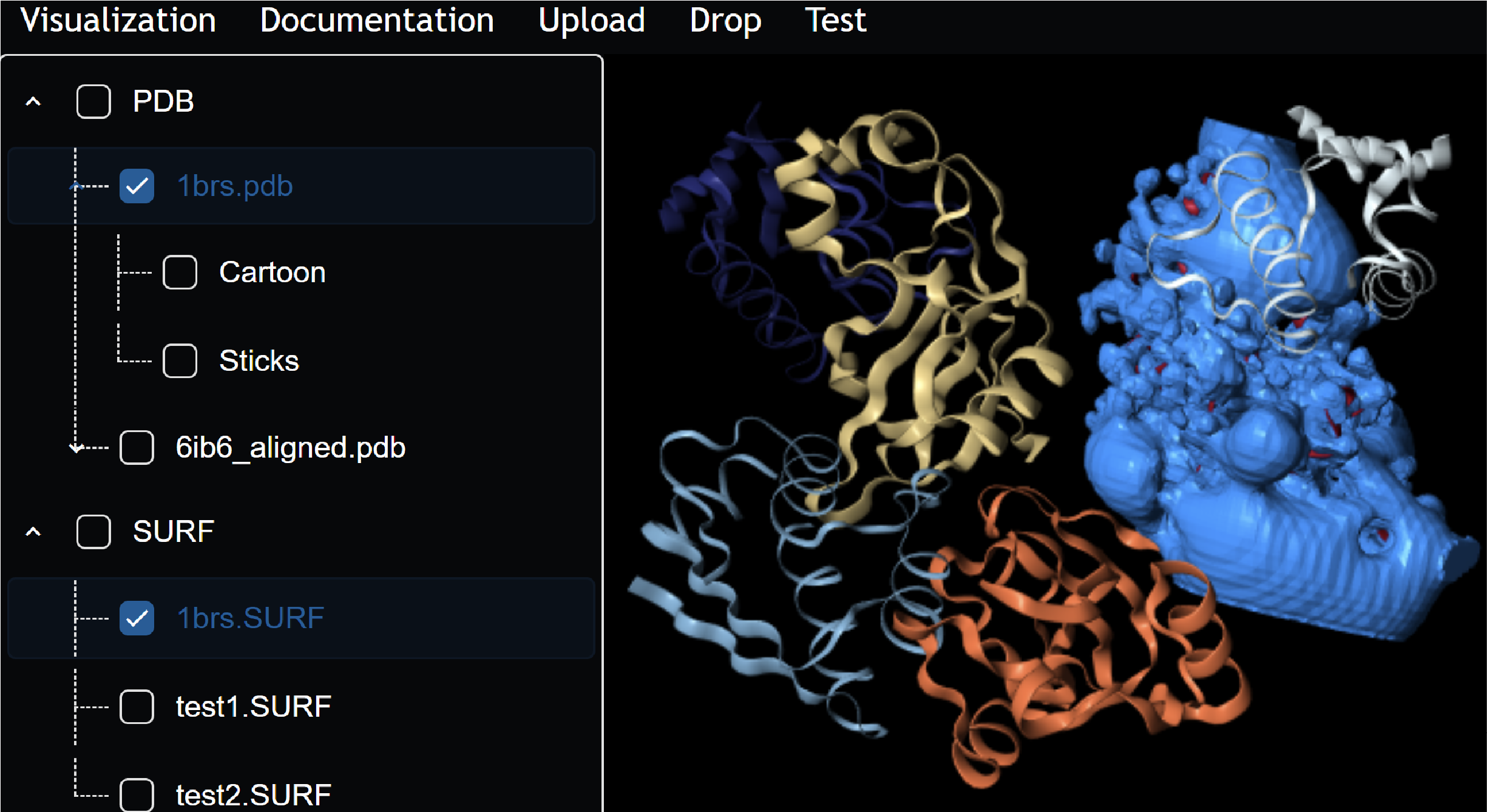
A Containerization Framework for Bioinformatics Software to Advance Scalability, Portability, and Maintainability.
In 14th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics (BCB '23), September 3–6, 2023
A full-stack web development project focused on the containerization and abstraction of Bioinformatics software to make web development easier for researchers. Awarded best paper finalist of CSBW at ACM-BCB '23'
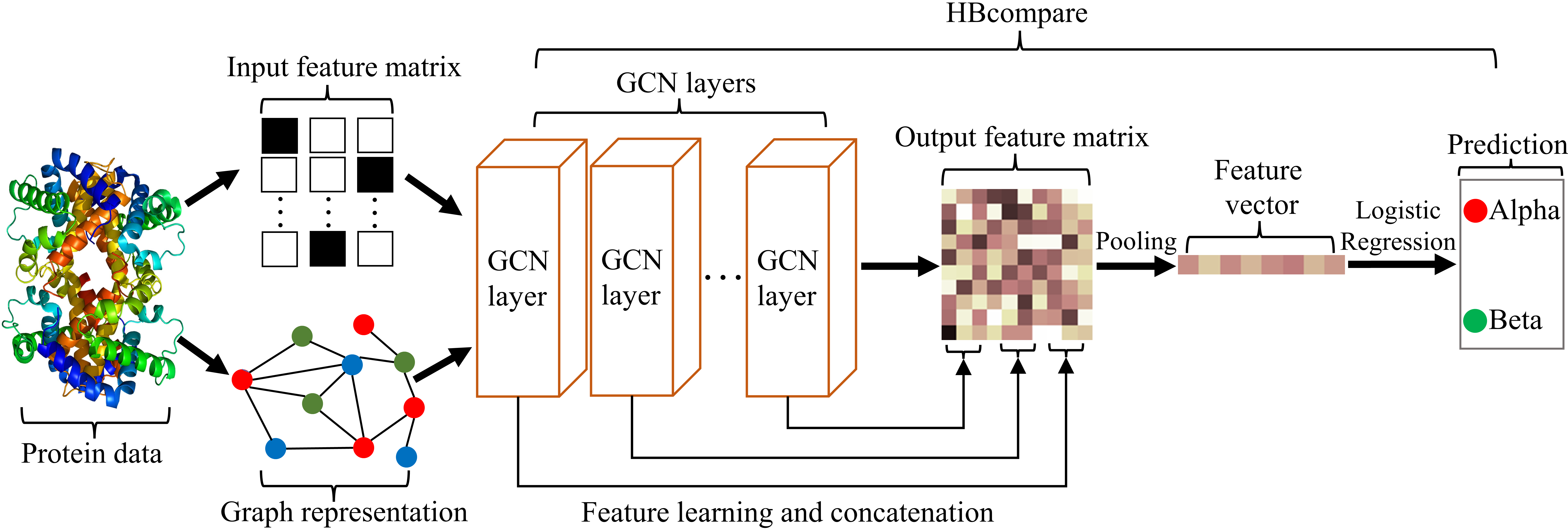
HBcompare: Classifying Ligand Binding Preferences with Hydrogen Bond Topology
Biomolecules, MDPI, Oct 28, 2022
A graph neural network approach to classifying hydrogen bond topology.
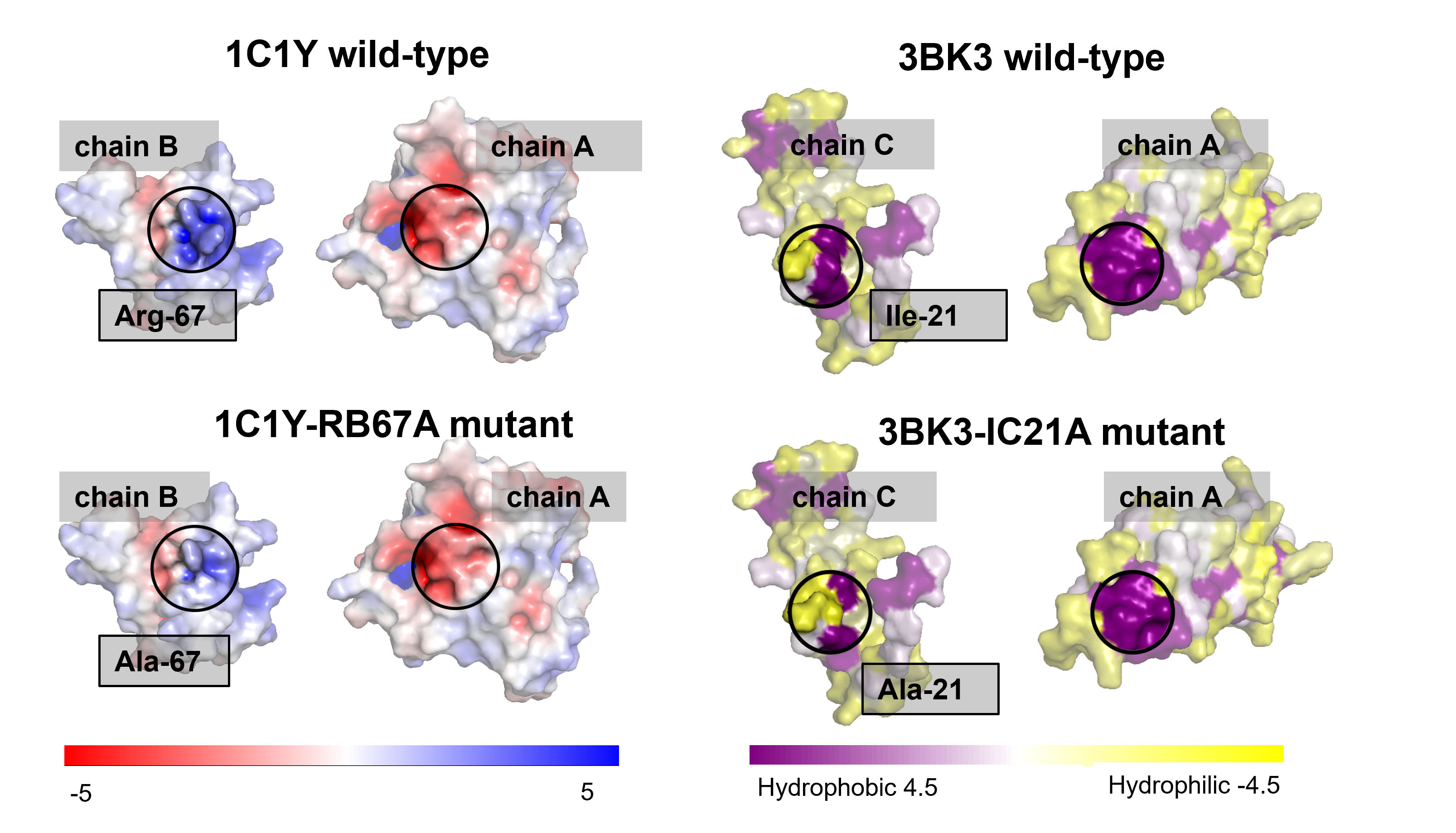
Analysis of Protein-Protein Interactions for Intermolecular Bond Prediction
Molecules, MDPI, Sep 21, 2022
Protein-Protein Interaction analysis using intermolecular bond prediction, mapped using coordinate structure and validated using the SKEMPI 2.0 data set.
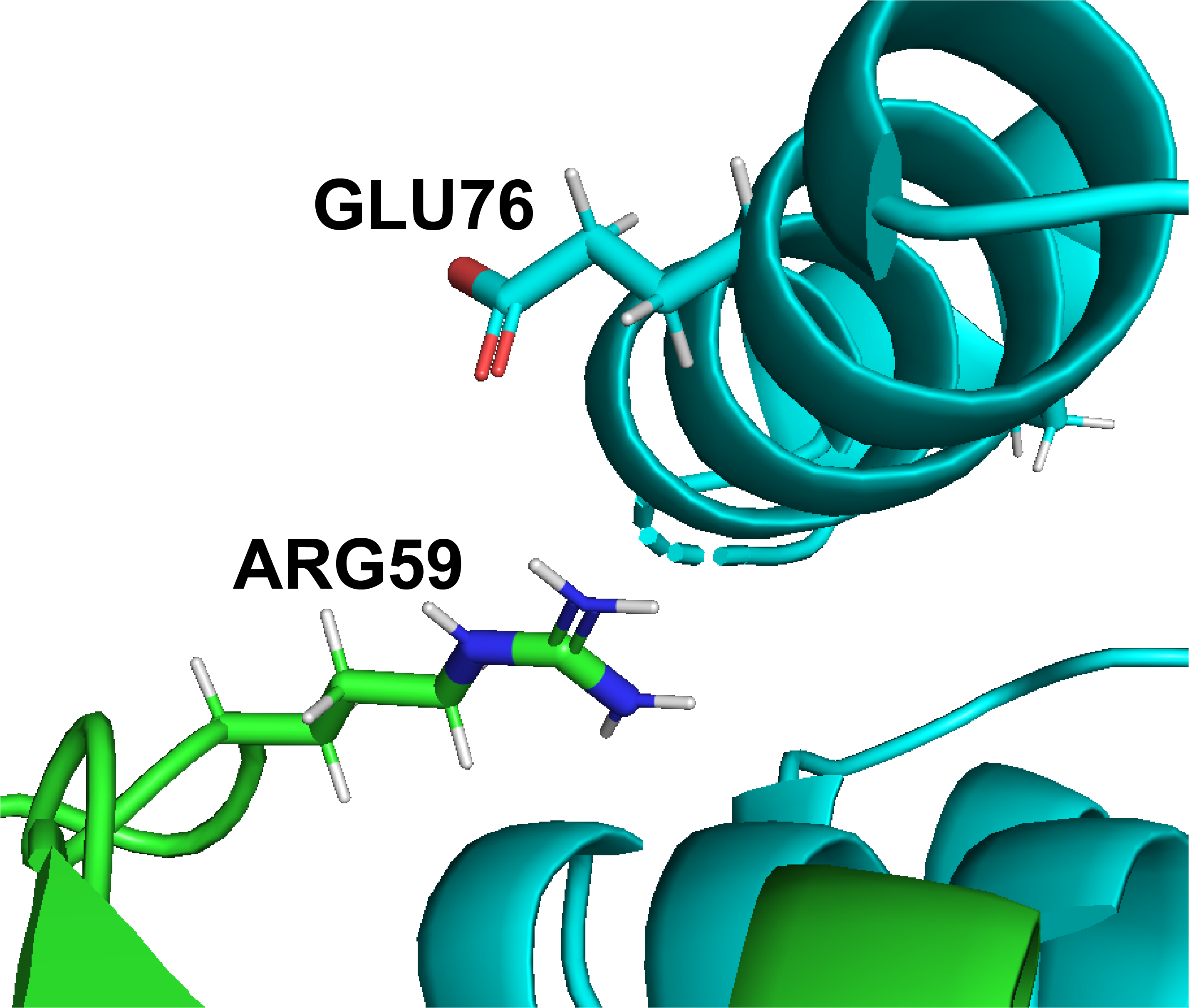
DiffBond: A Method for Predicting Intermolecular Bond Formation
2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE, Dec 9, 2021
An exploration of intermolecular bond topology in Protein-Protein Interactions.

Education
PhD Candidate in Computer Science/Bioinformatics
August 2020 - Graduating March 2026
Lehigh University, Bethlehem, PA
P.C. Rossin College of Engineering
Advisor: Brian Y. Chen
B.A. in Mathematics, Minor in Computer Science and Chemistry
August 2015 - May 2019
Skidmore College, Saratoga Springs, NY


